SAR problems: construct predictive theory.
Learn from chemical compounds of a known structure and activity.
3D molecular structures, large number of attributes: topological indices, molecular field parameters etc.
Finding most informative attributes - very important.
What do we look for?
- High accuracy.
- Explanatory power.
Neural and statistical methods have small explanatory power.
Inductive, rule discovery - usually better.
Continuous and quantized properties.
Pyrimidines and predictive toxicology evaluation
Two SAR problems:
- prediction of the antibiotic activity of pyrimidine compounds
- prediction of carcinogenicity of organic chemicals.
Pyrimidines
Class of chemical compounds with antibiotic activity.
They inhibit the activity of bacterial forms of some enzymes in a stronger way than the human forms and therefore kill bacteria.
A common template.
At three possible substitution positions chemical groups can be added.

3 substitution positions R3-R5.
Each chemical substitute has 9 features with a few symbolic attributes:
- group name,
- polarity, size,
- hydrogen-bond donor,
- hydrogen bond acceptor,
- pi-donor,
- pi-acceptor,
- polarizability,
- the sigma effect.
Pyrimidine template is described by 27 integer valued features.
No substitution - missing value, but very informative.
Pairs of chemicals (54 features) are compared.
Two classes: first compound has higher activity or vice versa.
2788 cases,
5-fold crossvalidation tests.
The predictive toxicology evaluation (PTE)
Oxford University Computing Laboratory challenge.
Based on US National Toxicology Program (NTP).
330 organic chemicals, 182 (55%) are carcinogenic, 148 non-carcinogenic.
417 features.
8 types of features:
- Features 1-69, atom type,
- feature 70, mutagenecity alert,
- features 71-285 so called WARMR alerts,
- no. 286-313 are counts of generic chemical groups found in the molecule,
- 314-376 are NTP bulk properties,
- 377-404 are various alerts that were used in \cite{ashby},
- 405-416 partial genotoxity test results,
- no. 417 is the AMES test.
- F. 418, class value: 0 non-carcinogenic, 1 carcinogenic, 2 unknown.
Test set include:
- 10 cases for which carcinogenicity is being experimentally determined.
- 20 other compounds of known carcinogenicity.
Large number of features, small number of test cases.
Statistical differences is not significant on test samples.
Better comparison using crossvalidation tests on all 350 known cases.
Feature Space Mapping neurofuzzy system
Theory for predicting structure relation activity is important.
Logical rules: decision trees, inductive logic programming, neural networks.
Feature Space Mapping (FSM) allows crisp and fuzzy rule sets.
FSM is a universal adaptive system.
Multidimensional separable functions modeling density of the input vectors.
Combinations of features define objects in the feature space, described by the joint density probability of the input/output data vectors using a network of properly parameterized transfer functions.
Gaussian type functions - the only radial separable functions:

X, input vector, D, the center of function, N(s), normalization factor.
Bicentral functions - soft rectangular membership functions:
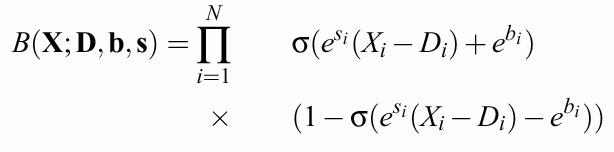
Logistic functions may be used here.
Adaptation: shifting the centers D, changing spreads b, rescaling slopes s.
Other localized separable functions: triangular, trapezoidal or rectangular functions:

Useful for extraction of crisp and fuzzy logical rules.
Separability facilitates interpretation - neurons provide "context-dependent" membership functions.
FSM is a neurofuzzy system, a density estimation network, a memory based system, a self-organizing system.
FSM architecture: network consists of three layers: an input, one hidden, and an output layer.
Conctructive algorithm: nodes added as needed.
Initial centers - clusterization using dendrograms or decision trees.
Dispersions and rotations of the clusters are optimized.
Output activation: probability, or confidence of the network in its classification.
FSM training algorithm: estimates probability density of input-output pairs in each class.
Results
Mean Spearman's rank correlation coefficient used:

n - number of pairs;
d - distance in rank of pairs;
-1<rs<1;
Mean Spearman's rank correlation coefficient for the pyrimidines dataset.
| Method | Rank correlation |
| CART | 0.499 |
| Linear Regression | 0.654 |
| Golem (ILP) | 0.684 |
| FSM | 0.780 |
Results from:
R.D. King, A. Srinivasan, M.J.E. Sternberg,
New Generation Computing (1995).
- Golem, ILP (inductive logic programming) system.
- FSM with Gaussian transfer functions - fuzzy rules.
High accuracy but low explonatory power.
- FSM, decision trees - large number of crisp rules.
PTE data: small number of training vectors, high dimensionality.
FSM with rectangle transfer function was done first.
60 features left out of 417 features, same used with Gaussian transfer functions.
Other algorithms:
- Distill Light - a stochastic algorithm,
- STEPS - evolutionary programming system,
- GloBo - stochastic system,
- OFAI - a combination of the two,
FSM results: Gaussians, optimization using crossvalidation on the training set.
FSM rules: 11 rules with 53 premises, using 24 features.
16 test vectors correctly (80%), 3 vectors unclassified, 1 error.
No correlation between results on the training/test set.
Accuracy on the test set: 1 case=5%.
FSM-rules: 1 error and 3 unknown cases.
| Method | Accuracy |
| Distill-Light | 90.0 |
| STEPS | 85.0 |
| GloBo | 85.0 |
| kNN, k=1, weighted | 80.0 |
| FSM-rules | 80.0 |
| FSM-Gauss | 75.0 |
| OFAI | 75.0 |
| Default | 70.0 |
Differences are not statistically significant.
Knowledge from the extracted rules may be useful.
C4.5rules, 10-fold crossvalidation - below the base rate were obtained, 53.6 ± 0.6%.
SSV decision tree: 62.0 ± 1.2 %.
FSM with Gaussians, 19 nodes, 64 ± 1.5% in 10xCV.
Initial clusterization is not significantly improved by learning.
Features are weak, with 40 best features 66% in the 10xCV on the training, 60-70% on the test is obtained after initial clusterization.
kNN, best results with k=1, Euclidean distance, feature selection.
16 features turned off, accuracy 63.2 ± 1.2% in 10xCV tests.
Minkowski's distance, large a=10, 40 features, 10xCV, accuracy 75.7 ± 0.7%, on test 75%.
Tuning scaling factors gives 77.7% on the training and 80% on the test data.
Conclusions
Pyrimidines: FSM results are significantly better but have little explanatory power.
Similar for other rule-based systems.
Alternative: good prototype cases, similarity based methods.
Toxicology data: hard to say.
Analysis of the rules by domain experts needed.
Use domain expert knowledge to pre-structure the FSM network.
Crossvalidation using several systems: accuracy higher than 80% using the features provided is unlikely.
Difficult problems, require further investigation.
Better features - difficult, requires quantum mechnical caluclations for all compounds.
Aggregation of features, combination of features?
SAR - good problems, requiring further development of methods.
Talks by Wlodzislaw Duch



